Introduction to ROOT
Overview
Teaching: min
Exercises: minQuestions
What is ROOT?
What can I learn here?
Objectives
Understand what is ROOT!
Introduction to ROOT!
What is ROOT?
Most likely some of you were already in touch with ROOT! ROOT is an open-source data analysis framework used by high energy physics and others, which lets you save and access your experiment’s data, allows you to process the data in a computationally efficient and statistically sound way and gives you access to all tools to produce publication-quality results. Basically, ROOT allows you to prcess data for physics analyses.
What can I learn here?
We would like to show you how you can perform efficient data analysis with ROOT! Starting with getting access to ROOT without any hassle, you will learn the advantages of ROOT in C++ and Python. Another ingredient for efficient analysis is a simple way to get help quickly and therefore you will learn where you can find support. ROOT can be used to go efficiently from the initial datasets to the result plots, all powered by ROOT!
Key Points
Understanding what is ROOT
Get ROOT
Overview
Teaching: min
Exercises: minQuestions
How to install ROOT on my system or get access to systems with ROOT pre-installed?
Objectives
Install ROOT
This section shows you multiple ways to get ROOT. Find below solutions to run ROOT locally, on LXPLUS, in batch systems and CIs!
Conda
Conda is a package management system and environment management system, which can install ROOT in a few minutes on Linux and MacOS.
The fastes way to get ROOT is installing Miniconda (a minimal Conda installation) and then run the following two commands:
conda create -c conda-forge --name <my-environment> root
conda activate <my-environment>
You can find detailed information about ROOT on Conda in this blog post.
CVMFS
CVMFS is a software distribution service that is already set up on many HEP systems. CMSSW including ROOT is distributed via CVMFS, but also other software stacks are available that contain ROOT.
Most notably, the CERN service LXPLUS has CVMFS always installed and enables rapid access to software and computing resources.
Docker
If you want to use ROOT in a CI system (e.g. GitLab pipelines or GitHub actions), most likely the software will be made available via Docker. The official ROOT docker containers can be found at https://hub.docker.com/r/rootproject/root. The different base images and ROOT versions are encoded in the tags, for example 6.22.00-ubuntu20.04, and latest will get you the latest ROOT release (v6.22) based on Ubuntu 20.04. If you want to try it, get Docker and run the following command to start the container with a bash shell.
docker run --rm -it rootproject/root /bin/bash
Binary releases and packages
The classic way to distribute software, besides the source code, are plain binary releases. You can download these from the release pages on https://root.cern/install/all_releases for all major MacOS and Linux versions. If you choose this installation method, make sure ROOT dependencies are installed on your system. Complete installation instructions for binary releases are available here.
In addition, for some Linux distributions, the ROOT community maintains packages in the respective package managers. You can find a list of maintained packages at https://root.cern/install/#linux-package-managers.
Verify the ROOT version
Since ROOT has a long history and numerous releases, on old systems such Scientific Linux 6 you may find correspondingly old ROOT versions. However, with the following commands you can easily verify your ROOT version and also find expert details about the ROOT configuration!
# ROOT version and build tag
root --version
# Again the ROOT version (this also works with older ROOT versions)
root-config --version
# Check that ROOT was built with C++14 support
# The output must contain one of -std=c++{14,17,1z} so that all code examples of this lesson run!
root-config --cflags
# List all the ROOT configuration options that can be checked
root-config --help
Find your way to access ROOT!
For the exercises later you need at least ROOT 6.18 and C++14 support. Feel free to set up for yourself your preferred environment satisfying this requirement!
Fallback solution
As a fallback solution you can always connect to LXPLUS via
ssh -Y your_username@lxplus.cern.ch. The-Yflag enables X forwarding, which allows you to forward the output of graphics application in case you run a system with an X server such as almost all Linux distributions.
Key Points
ROOT is accessible in several different ways
Detailed up-to-date instructions can be found at https://root.cern/install
ROOT and using ROOT prompt
Overview
Teaching: min
Exercises: minQuestions
How to use a ROOT file and the ROOT prompt ?
Objectives
Open a ROOT file
Use ROOT prompt
Interactive C++
One of the main features of ROOT is the possibility to use C++ interactively thanks to the C++ interpreter Cling. Cling lets you use C++ just like Python either from the prompt or in scripts.
The ROOT prompt
By just typing root in the terminal you will enter the ROOT prompt. Like the Python prompt, the ROOT prompt is well suited to fast investigations.
$ root
root [0] 1+1
(int) 2
If you pass a file as argument to root, the file will be opened when entering the prompt and put in the variable _file0. ROOT typically comes with support for reading files remotely via HTTP (and XRootD), which we will use for the following example:
No support for remote files?
Although unlikely, your ROOT build may not be configured to support remote file access. In this case, you can just download the file with
curl -O https://root.cern/files/tmva_class_example.rootand point to your local file. No other changes required!
$ root tmva_class_example.root
root [0]
Attaching file mva_class_example.root as _file0...
(TFile *) 0x555f82beca10
root [1] _file0->ls() // Show content of the file, all objects are accessible via the prompt!
TWebFile** https://root.cern/files/tmva_class_example.root
TWebFile* https://root.cern/files/tmva_class_example.root
KEY: TTree TreeS;1 TreeS
KEY: TTree TreeB;1 TreeB
root [2] TreeS->GetEntries() // Number of events in the dataset
root [3] TreeS->Print() // Show dataset structure
******************************************************************************
*Tree :TreeS : TreeS *
*Entries : 6000 : Total = 98896 bytes File Size = 89768 *
* : : Tree compression factor = 1.00 *
******************************************************************************
*Br 0 :var1 : var1/F *
*Entries : 6000 : Total Size= 24641 bytes One basket in memory *
*Baskets : 0 : Basket Size= 32000 bytes Compression= 1.00 *
*............................................................................*
*Br 1 :var2 : var2/F *
*Entries : 6000 : Total Size= 24641 bytes One basket in memory *
*Baskets : 0 : Basket Size= 32000 bytes Compression= 1.00 *
*............................................................................*
*Br 2 :var3 : var3/F *
*Entries : 6000 : Total Size= 24641 bytes One basket in memory *
*Baskets : 0 : Basket Size= 32000 bytes Compression= 1.00 *
*............................................................................*
*Br 3 :var4 : var4/F *
*Entries : 6000 : Total Size= 24641 bytes One basket in memory *
*Baskets : 0 : Basket Size= 32000 bytes Compression= 1.00 *
*............................................................................*
root [4] TreeS->Draw("var1") // Draw a histogram of the variable var1
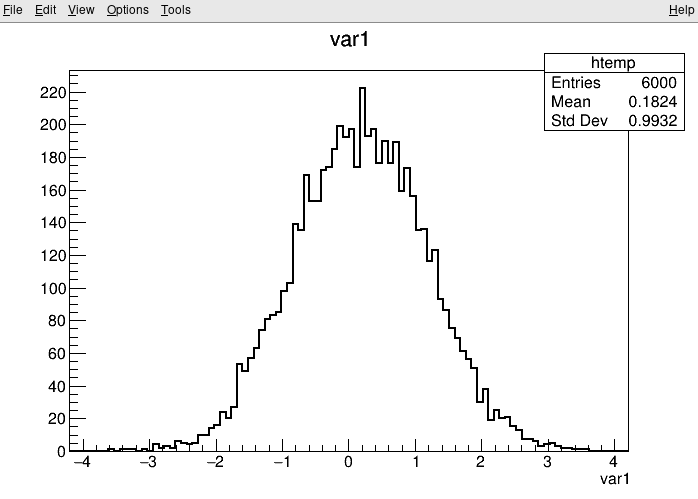
Investigating data in ROOT files
You have already seen the usage of TTree::Draw in the previous section. Such quick investigations of data in ROOT files are typical usecases which most analysts encounter on a daily basis. In the following you can learn about different ways to approach this task!
Manually plotting with TTree::Draw
For quick studies on the raw data in a TTree on the command line, you can use TTree::Draw to make simple visualizations:
$ root https://root.cern/files/tmva_class_example.root
root [0]
Attaching file https://root.cern/files/tmva_class_example.root as _file0...
(TFile *) 0x558d7b54aa50
root [1] TreeS->Draw("var1") // just draw var1
Info in <TCanvas::MakeDefCanvas>: created default TCanvas with name c1
root [2] TreeS->Draw("var1", "var2 > var1", "SAME") // draw var1 with the selection var2 > var1
(long long) 3222
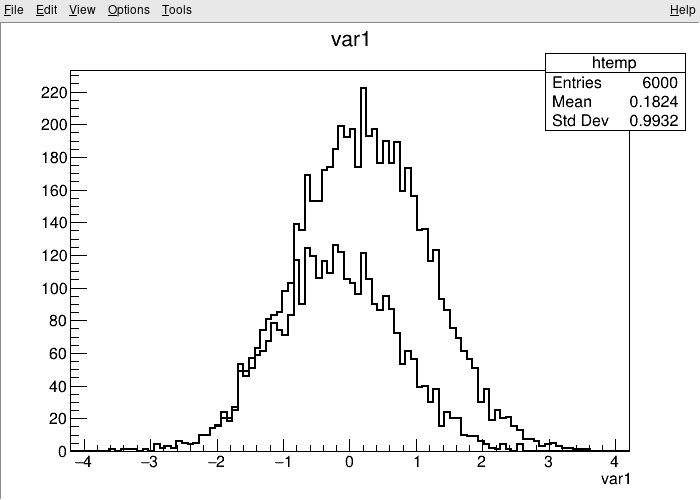
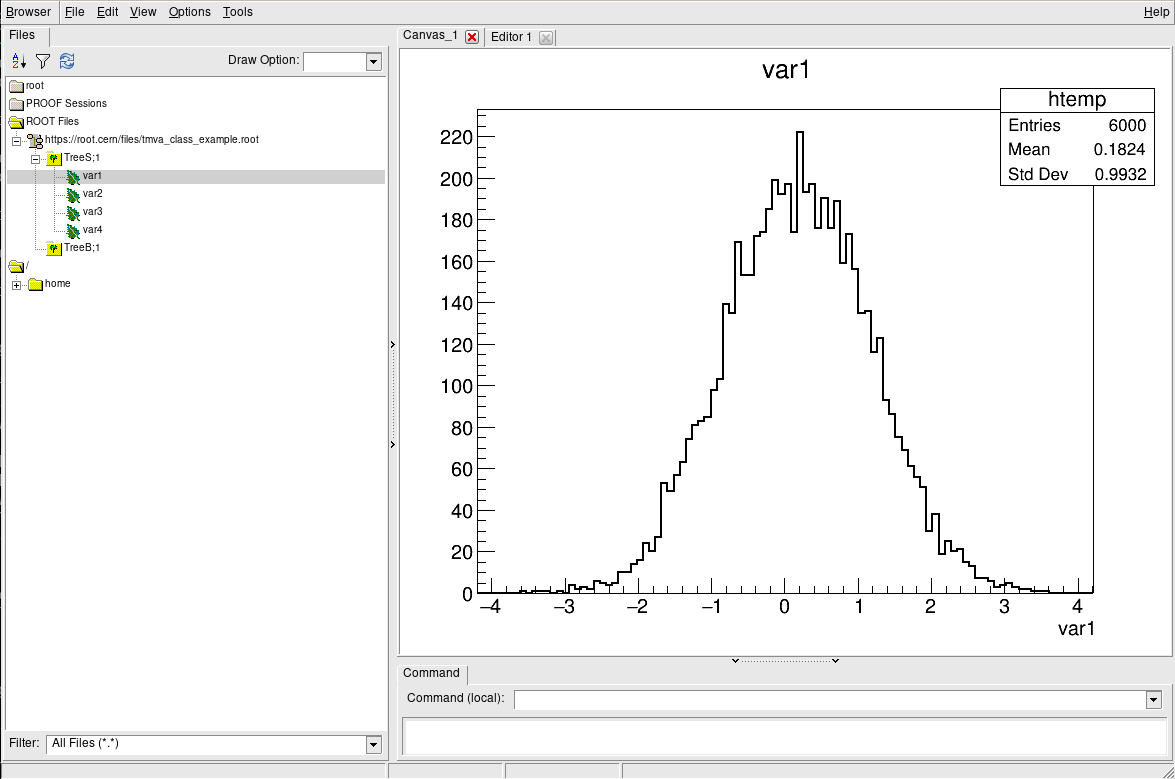
The TBrowser
More convenient is using ROOT’s tool for browsing ROOT files, the TBrowser. You can spawn the GUI directly from the ROOT prompt as shown below.
$ root https://root.cern/files/tmva_class_example.root
root [0]
Attaching file https://root.cern/files/tmva_class_example.root as _file0...
(TFile *) 0x557892a0ef10
root [1] TBrowser b
(TBrowser &) Name: Browser Title: ROOT Object Browser
The rootbrowse executable
For convenience, ROOT provides the executable rootbrowse, which lets you open a TBrowser directly from the command line and display the files given as arguments!
$ rootbrowse https://root.cern/files/tmva_class_example.root
Other ROOT executables
There are many small helpers shipped with ROOT, which let you operate on data quickly from the command line and solve typical day-to-day tasks with ROOT files.
List of ROOT executables
rootbrowse: Open a ROOT file and a TBrowserrootls: List file content, tree branches, objects’ statsrootcp: Copy objects within a file or between filesrootdrawtree: Simple analyses from the command linerooteventselector: Select branches, events, compression algorithms and extract slimmer treesrootmkdir: Creates a directory in a TFilerootmv: Move objects between filesrootprint: Print objects in plots on filesrootrm: Remove objects from files
$ rootls https://root.cern/files/tmva_class_example.root
TreeB TreeS
$ rootls -l https://root.cern/files/tmva_class_example.root
TTree Jan 19 14:25 2009 TreeB "TreeB"
TTree Jan 19 14:25 2009 TreeS "TreeS"
$ rootls -t https://root.cern/files/tmva_class_example.root
TTree Jan 19 14:25 2009 TreeB "TreeB"
var1 "var1/F" 0
var2 "var2/F" 0
var3 "var3/F" 0
var4 "var4/F" 0
weight "weight/F" 0
Cluster INCLUSIVE ranges:
- # 0: [0, 5998]
- # 1: [5999, 5999]
The total number of clusters is 2
TTree Jan 19 14:25 2009 TreeS "TreeS"
var1 "var1/F" 0
var2 "var2/F" 0
var3 "var3/F" 0
var4 "var4/F" 0
Cluster INCLUSIVE ranges:
- # 0: [0, 5998]
- # 1: [5999, 5999]
The total number of clusters is 2
Try it by yourself!
Feel free to investigate the tools presented here!
Key Points
Learn hoe to open a ROOT file.
Learn to use the ROOT prompt.
ROOT scripts
Overview
Teaching: min
Exercises: minQuestions
How to use ROOT scripts?
Objectives
Learn about using C++ scripts with ROOT
This section is dedicated to the introduction to using ROOT scripts or macros.
ROOT scripts
A unique feature of ROOT is the possibility to use C++ scripts, also called “ROOT macros”. A ROOT script contains valid C++ code and uses as entrypoint a function with the same name as the script. Let’s take as example the file myScript.C with the following content.
void myScript() {
auto file = TFile::Open("tmva_class_example.root");
auto newtree = (TTree*)file->Get("TreeS");
auto entries = newtree->GetEntries();
std::cout << "Entried in TreeS : " << entries << std::endl;
}
Scripts can be processed by passing them as argument to the root executable:
$ root myScript.C
root [0]
Processing myScript.C...
Entried in TreeS : 6000
There are other ways to run scripts, try them.
Plotting from file ith script
void myScript_plot() {
TH1F *h_var1 = new TH1F("h_var1", "var1", 100, -4.0, 4.0);
TFile *file = new TFile("/Users/sar/Desktop/TestROOT/tmva_class_example.root");
TTree *tree1 = (TTree*)file->Get("TreeS");
float var1;
tree1->SetBranchAddress("var1", &var1);
int nentries = (int)tree1->GetEntries();
for(int i=0; i < nentries; i++){
tree1->GetEntry(i);
h_var1->Fill(var1);
}
TCanvas *c1 = new TCanvas();
h_var1->Draw();
}
Examples of creating histogam:
void scriptHist(){
auto cnt_r_h=new TH1F("count_rate",
"Count Rate;N_{Counts};# occurencies",
100, // Number of Bins
-0.5, // Lower X Boundary
15.5); // Upper X Boundary
auto mean_count=3.6f;
TRandom3 rndgen;
// simulate the measurements
for (int imeas=0;imeas<400;imeas++)
cnt_r_h->Fill(rndgen.Poisson(mean_count));
auto c= new TCanvas();
cnt_r_h->Draw();
auto c_norm= new TCanvas();
cnt_r_h->DrawNormalized();
// Print summary
cout << "Moments of Distribution:\n"
<< " - Mean = " << cnt_r_h->GetMean() << " +- "
<< cnt_r_h->GetMeanError() << "\n"
<< " - Std Dev = " << cnt_r_h->GetStdDev() << " +- "
<< cnt_r_h->GetStdDevError() << "\n"
<< " - Skewness = " << cnt_r_h->GetSkewness() << "\n"
<< " - Kurtosis = " << cnt_r_h->GetKurtosis() << "\n";
}
Using graphs
// Builds a graph with errors, displays it and saves it as
// image. First, include some header files
// (not necessary for Cling)
#include "TCanvas.h"
#include "TROOT.h"
#include "TGraphErrors.h"
#include "TF1.h"
#include "TLegend.h"
#include "TArrow.h"
#include "TLatex.h"
void macro1(){
// The values and the errors on the Y axis
const int n_points=10;
double x_vals[n_points]=
{1,2,3,4,5,6,7,8,9,10};
double y_vals[n_points]=
{6,12,14,20,22,24,35,45,44,53};
double y_errs[n_points]=
{5,5,4.7,4.5,4.2,5.1,2.9,4.1,4.8,5.43};
// Instance of the graph
TGraphErrors graph(n_points,x_vals,y_vals,nullptr,y_errs);
graph.SetTitle("Measurement XYZ;lenght [cm];Arb.Units");
// Make the plot estetically better
graph.SetMarkerStyle(kOpenCircle);
graph.SetMarkerColor(kBlue);
graph.SetLineColor(kBlue);
// The canvas on which we'll draw the graph
auto mycanvas = new TCanvas();
// Draw the graph !
graph.DrawClone("APE");
// Define a linear function
TF1 f("Linear law","[0]+x*[1]",.5,10.5);
// Let's make the function line nicer
f.SetLineColor(kRed); f.SetLineStyle(2);
// Fit it to the graph and draw it
graph.Fit(&f);
f.DrawClone("Same");
// Build and Draw a legend
TLegend leg(.1,.7,.3,.9,"Lab. Lesson 1");
leg.SetFillColor(0);
graph.SetFillColor(0);
leg.AddEntry(&graph,"Exp. Points");
leg.AddEntry(&f,"Th. Law");
leg.DrawClone("Same");
// Draw an arrow on the canvas
TArrow arrow(8,8,6.2,23,0.02,"|>");
arrow.SetLineWidth(2);
arrow.DrawClone();
// Add some text to the plot
TLatex text(8.2,7.5,"#splitline{Maximum}{Deviation}");
text.DrawClone();
mycanvas->Print("graph_with_law.pdf");
}
int main(){
macro1();
}
Examples of creating a ROOT file:
void myScript_file() {
TH1F *h_var1 = new TH1F("h_var1", "var1", 100, -4.0, 4.0);
TFile *file = new TFile("tmva_class_example.root");
TTree *tree1 = (TTree*)file->Get("TreeS");
float varx;
tree1->SetBranchAddress("var1", &varx);
TFile* newFile( TFile::Open("newfile.root", "RECREATE") );
auto newtree = std::make_unique<TTree>("newtree", "The Tree Title");
newtree->Branch("branch0", &varx);
int nentries = (int)tree1->GetEntries();
for(int i=0; i < nentries; i++){
tree1->GetEntry(i);
h_var1->Fill(varx);
newtree->Fill();
}
TCanvas *c1 = new TCanvas();
h_var1->Draw();
newFile->cd();
newtree->Write();
h_var1->Write();
newFile->Write();
newFile->Close();
}
The advantage of such scripts is the simple interaction with C++ libraries (such as ROOT) and running your code at C++ speed with the convenience of a script.
Compiled C++
You can improve the runtime of your programs if you compile them upfront. Therefore, ROOT tries to make the compilation of ROOT macros as convenient as possible!
ACLiC
ROOT provides a mechanism called ACLiC to compile the script in a shared library and call the compiled code from interactive C++, all automatically!
The only change required to our script is that we need to include all required headers:
#include "TFile.h"
#include "TTree.h"
#include <iostream>
void myScript() {
// The body of the myScript function goes here
}
Now, let’s compile and run the script again. Note the + after the script name!
$ root myScript.C+
ACLiC has many more features, for example compiling your program with debug symbols using +g. You can find the documentation here.
C++ compilers
Of course, the C++ code can also just be compiled with C++ compilers such as g++ or clang++ with the advantage that you have full control of all compiler settings, most notable the optimization flags such as -O3!
To do so, we have to add the main function to the script, which is the default entrypoint for C(++) programs.
#include "TFile.h"
#include "TTree.h"
#include <iostream>
void myScript() {
// The body of the myScript function goes here
}
int main() {
myScript();
return 0;
}
Now, you can use the following command with your C++ compiler of choice to compile the script into an executable.
$ g++ -O3 -o myScript myScript.C $(root-config --cflags --libs)
$ ./myScript
TreeS : 6000
TreeB : 6000
Computationally heavy programs and long running analyses may benefit greatly from the optimized compilation with -O3 and can save you hours of computing time!
Key Points
ROOT provides many features from histogramming, fitting and plotting to investigating data interactively in C++ and Python but scripts can help with advanced work
Learn about C/C++ scripts for ROOT.
Using ROOT with Python
Overview
Teaching: min
Exercises: minQuestions
How can I use ROOT with Python?
Objectives
Learn about the basics of ROOT with Python
Python
ROOT provides the Python bindings called PyROOT. PyROOT is not just ROOT from Python, but a full-featured interface to call C++ libraries in a pythonic way. This lets you import the ROOT module from Python and makes all features dynamically available. Let’s rewrite the C++ example from above and put the code in the file myScript.py!
import ROOT
rfile = ROOT.TFile.Open('https://root.cern/files/tmva_class_example.root')
for key in rfile.GetListOfKeys():
name = key.GetName()
entries = rfile.Get(name).GetEntries()
print('{} : {}'.format(name, entries))
Calling the Python script works as expected:
$ python myScript.py
TreeS : 6000
TreeB : 6000
But PyROOT can do much more for you than simply providing access to C++ libraries from Python. You can also inject efficient C++ code into your Python program to speed up potentially slow parts of your program!
import ROOT
ROOT.gInterpreter.Declare('''
int my_heavy_computation(std::string x) {
// heavy computation goes here
return x.length();
}
''')
# Functions and object made available via the interpreter are accessible from
# the ROOT module
y = ROOT.my_heavy_computation("the ultimate answer to life and everything")
print(y) # Guess the result!
Basic histogramming, fitting and plotting
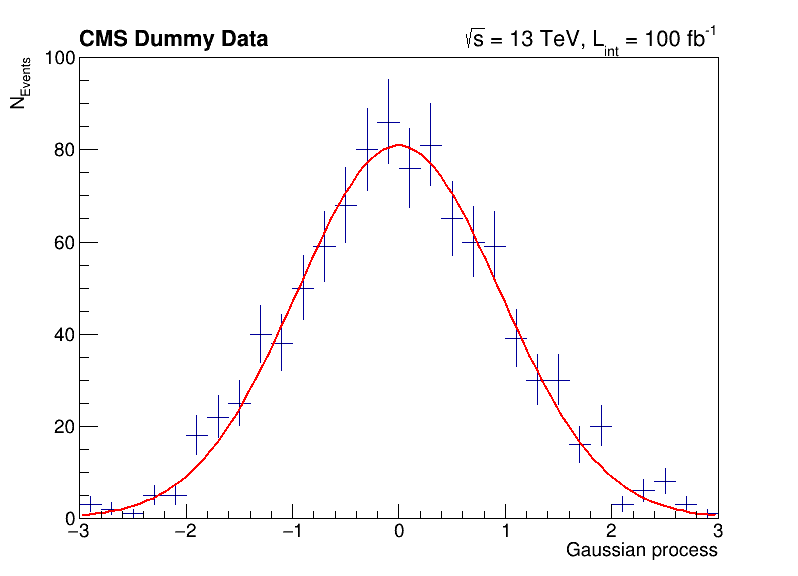
The following script uses basic features from ROOT, which are used commonly in day-to-day work with ROOT. You can investigate the typical workflow to create histograms with TH1F, fit a function to the data with TF1 and produce an accurate visualization with TCanvas and others. Below, you can see the output of the fit to the data with the measured parameters.
import ROOT
import numpy as np
# Make global style changes
ROOT.gStyle.SetOptStat(0) # Disable the statistics box
ROOT.gStyle.SetTextFont(42)
# Create a canvas
c = ROOT.TCanvas('c', 'my canvas', 800, 600)
# Create a histogram with some dummy data and draw it
data = np.random.randn(1000).astype(np.float32)
h = ROOT.TH1F('h', ';Gaussian process; N_{Events}', 30, -3, 3)
for x in data: h.Fill(x)
h.Draw('E')
# Fit a Gaussian function to the data
f = ROOT.TF1('f', '[0] * exp(-0.5 * ((x - [1]) / [2])**2)')
f.SetParameters(100, 0, 1)
h.Fit(f)
# Let's add some CMS style headline
label = ROOT.TLatex()
label.SetNDC(True)
label.SetTextSize(0.040)
label.DrawLatex(0.10, 0.92, '#bf{CMS Dummy Data}')
label.DrawLatex(0.58, 0.92, '#sqrt{s} = 13 TeV, L_{int} = 100 fb^{-1}')
# Save as png file and show interactively
c.SaveAs('dummy_data.png')
c.Draw()
FCN=30.2937 FROM MIGRAD STATUS=CONVERGED 67 CALLS 68 TOTAL
EDM=1.34686e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 p0 8.09397e+01 3.19887e+00 7.10307e-03 -3.40988e-05
2 p1 -3.46483e-03 3.10501e-02 8.47265e-05 -2.30742e-03
3 p2 9.56532e-01 2.24141e-02 4.97399e-05 2.58872e-03
Info in <TCanvas::Print>: file dummy_data.png has been created
Try it by yourself!
Run the example code by yourself! In case the execution ends without displaying the plot on screen, you can run the script in interpreted mode with
python -i your_script.py. That will keep the process alive after the plot is displayed.
A guide to such advanced features of PyROOT can be found in the official manual at https://root.cern/manual/python. Feel free to investigate!
Interoperability with NumPy arrays
There are many reasons, for example machine learning applications, to want to export your data in Python to NumPy arrays. This is easily possible with ROOT and is part of RDataFrame. The code snippets below show you how to do this conversion and how to move the data to typical tools in the Python ecosystem, e.g., numpy and pandas.
numpy and pandas
Have you installed
numpyandpandasor are you on a system which has them available? Normally, you can just runpip install --user numpy pandasto install missing packages! Another option is searching in your system package manager, they are typically available on all platforms.
Convert data in ROOT files to numpy arrays
The conversion feature is attached to the class RDataFrame. We will not introduce you here to this way to process data with ROOT because the following section is dedicated to RDataFrame. For now, just keep in mind that you call AsNumpy! The data is returned as a dictionary of one-dimensional numpy arrays.
# Read out the data as a dictionary of numpy arrays
import ROOT
df = ROOT.RDataFrame('TreeS', 'https://root.cern/files/tmva_class_example.root')
columns = ['var1', 'var2', 'var3', 'var4']
data = df.AsNumpy(columns)
print('var1: {}'.format(data['var1']))
var1: [-1.1436108 2.1434433 -0.44391322 ... 0.37746507 -2.072639 -0.09141494]
Move the data to numpy or pandas
The data can be passed naturally to any method in the Python ecosystem which processes numpy arrays. Below is an example that computes the mean of each column.
# Apply numpy methods
import numpy as np
print('Means: {}'.format([np.mean(data[c]).item() for c in columns]))
Means: [0.18244409561157227, 0.28425973653793335, 0.3789360225200653, 0.7712161540985107]
Another interesting usecase is moving the dataset directly to a pandas dataframe. You can use the output of AsNumpy directly as input to its constructor.
# Convert to a pandas dataframe
import pandas
pdf = pandas.DataFrame(data)
print(pdf)
var1 var2 var3 var4
0 -1.143611 -0.822373 -0.495426 -0.629427
1 2.143443 -0.018923 0.267030 1.267493
2 -0.443913 0.486827 0.139535 0.611483
3 0.281100 -0.347094 -0.240525 0.347208
4 0.604006 0.151232 0.964091 1.227711
... ... ... ... ...
5995 -0.040650 -0.154212 -0.097715 0.440331
5996 0.099931 -1.183759 0.034616 0.644502
5997 0.377465 -0.030945 1.166082 0.728614
5998 -2.072639 -0.635586 -0.747371 -1.285679
5999 -0.091415 0.221271 0.569032 1.386130
[6000 rows x 4 columns]
Try it by yourself!
The statements are very short, you can just copy paste them into the Python prompt. Feel free to investigate what you can do with
AsNumpy! Further information can be found here.
ROOT in Jupyter notebooks
ROOT provides a deep integration with Jupyter notebooks. You can start a Jupyter notebook server including ROOT features with the following command:
root --notebook
Alternatively, you can go to https://swan.cern.ch, which provides Jupyter notebooks integrated with CERN’s cloud storage as a web service. Note that you may have to visit https://cernbox.cern.ch first at least once with your user account to create your CERNBox space!
Python kernel
Jupyter is often use to edit Python code interactively. By creating a new notebook with a Python kernel, you will see something similar to the screenshot below and you can work interactively with Python in the browser!
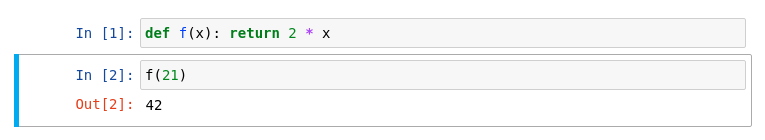
C++ kernel
ROOT provides a Jupyter C++ kernel, which behaves similarly to the Python kernel but for C++! Similar to the ROOT prompt, you can work interactively with C++ in the notebook. Just select the C++ kernel in the drop-down menu!
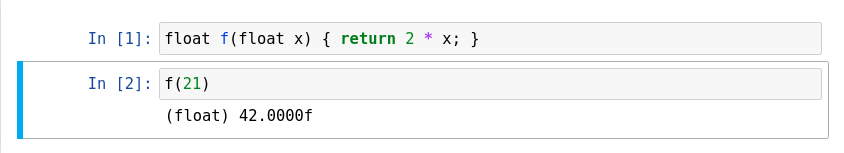
JSROOT
Another feature of ROOT is the %jsroot on magic, which enables ROOT’s JavaScript integration! This allows you to interact with the visualization such as you are used to it from the interactive graphics in the Python prompt.
Because it’s JavaScript, we can also embed these plots easily in any website. You can find an interactive version of the plot from the top of this section at the bottom of the page. For example, you can zoom in, add grid lines or get detailed information about the data points, right here!
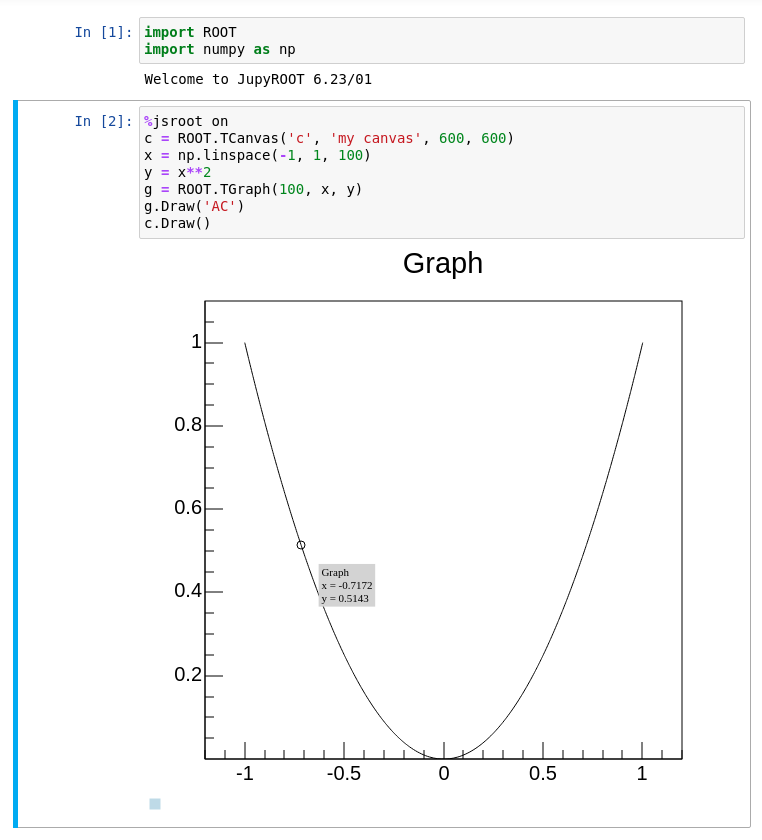
Try it by yourself!
Either run Jupyter locally via
root --notebookor go to https://swan.cern.ch to try ROOT in a Jupyter notebook!
More useful features
ROOT is made for HEP analysis and contains many other features that are useful in typical tasks, for example:
- TEfficiency, to handle histograms representing efficiencies and their uncertainties
- THStack, to stack histograms
- TRatioPlot, to create ratio plots with the histograms on the top and the ratio with the correct uncertainties on the bottom
- And many more!
Try using ROOT with interactive C++, compiled C++ and Python!
Make yourself familiar with the different ways you can run an analysis with ROOT!
Key Points
ROOT can be used with python
PyROOT lets you use C++ from Python but offers many more advanced features to speed up your analysis in Python. Details about the dynamic Python bindings provided by PyROOT can be found on https://root.cern/manual/python.
How to get help with ROOT?
Overview
Teaching: min
Exercises: minQuestions
Where can I find documentation?
Where can I ask for help?
Objectives
Learn how to find the official documentation!
Learn about the ROOT forum to get help!
Something does not work as expected, how can I get help?
User support is an integral part of ROOT and happily provided by the ROOT team!
We provide multiple communication channels so that we can help you but also you can help yourself to find the right answers to your questions, as fast as possible!
Support and discussion channel for this lesson
Communication channels for support and discussion dedicated to this lesson are linked on the front page!
The ROOT website, the beginner’s guide and the manual
The ROOT website is home to the beginner’s guide and the more in-depth manual. These are a great resource to start with ROOT and learn about parts of the framework in high detail. Keep in mind the ROOT website at https://root.cern, which provides links to all resources in a single place!
The reference guide
The reference guide provides a more technical documentation about ROOT powered by Doxygen. You can search for classes or functions in ROOT, learn about types and methods and trace features down to the actual implementation.
Although the reference guide is more technical in first place, important classes have extensive additional documentation. Feel free to investigate TTree or RDataFrame!
Another part of the reference guide are the tutorials, which explain features in working code examples. Feel free to look at tutorials for RooFit and RDataFrame, which cover many typical use cases for these parts of ROOT!
The ROOT forum
The ROOT forum is the to-go place if you cannot find the answer in the documentation. Don’t hesitate to open a discussion, there is always someone from the ROOT team actively taking care of new questions in the forum!
But not only questions are very welcome, you can also discuss possible improvements or make suggestions for new features!
Bug tracking
Bugs are currently tracked on Jira, but we will soon switch to GitHub issues. However, if you discover bugs, please report them! In case you are not sure whether you see a bug or a feature, posting in the ROOT forum is always a good idea and always appreciated!
Key Points
User support is an integral part of ROOT!
https://root.cern is the entry point to find all documentation
The reference guide provides in-depth technical documentation, but also additional explanation for classes and a huge amount of tutorials explaining features with code
The ROOT forum is actively maintained by the ROOT team to support you!
Assignment 1 Hints
Overview
Teaching: min
Exercises: minQuestions
How to complete Assignment 1 ?
Objectives
Complete Assignment 1
Read and write file with arrays as branches and calculating invariant mass
The following script should help you understand how to read and write a file with arrays as branches, and some hints on calculating invariant mass of a particle that decays to two objects which are reconstructed. Do read the comments embedded within the code.
void myScript() {
auto inputfile = TFile::Open("DYJetsToLL.root");
auto inputtree = (TTree*)inputfile->Get("Events");
long nentries = inputtree->GetEntries();
const int nmuonmax = 50;// a number higher than maximum number of muons in any event in the file
const int ntaumax = 50;// a number higher than maximum number of taus in any event in the file
uint nmuon;
uint ntau;
float taupt[ntaumax];//these are arrays, that is they contain multiple numbers
float tauphi[ntaumax];
float taueta[ntaumax];
float taumass[ntaumax];
int taucharge[ntaumax];
bool tauisotight[ntaumax]; //this is a boolean array
TLorentzVector lvectau1, lvectau2, lvecZ;//Lorentz vectors
float ditaumass;
//Setting branches of input file and tree to the variables
inputtree->SetBranchAddress("nMuon", &nmuon);
inputtree->SetBranchAddress("nTau", &ntau);
inputtree->SetBranchAddress("Tau_pt", taupt); //syntax for assigning branches to arrays
inputtree->SetBranchAddress("Tau_phi", tauphi);
inputtree->SetBranchAddress("Tau_eta", taueta);
inputtree->SetBranchAddress("Tau_charge", taucharge);
inputtree->SetBranchAddress("Tau_mass", taumass);
inputtree->SetBranchAddress("Tau_idIsoTight", tauisotight); //this is a boolean array
//Output file
TFile* newFile( TFile::Open("newfile.root", "RECREATE") );
auto newtree = std::make_unique<TTree>("newtree", "Events");
newtree->Branch("nTau", &ntau, "nTau/I");//no need for the "nTau/I" as it takes it as default, but specifying for following cases
newtree->Branch("Tau_pt", taupt, "Tau_pt[nTau]/F");//for arrays, need to specify in the third argument that it is an array (notice, no & sign in second arguments for array) of size ntau and store float variable
//Output histograms
auto hntau = new TH1F("hntau", "Number of Taus", 50, -0.5, 49.5);
auto htaupt = new TH1F("htaupt", "Tau pT", 100, 0.0, 200.0);
//Event loop
std::cout<<"Entering event loop"<<std::endl;
for (int i = 0; i < int(nentries); i++){
inputtree->GetEntry(i);
hntau->Fill(ntau);
for (auto j = 0; j < ntau; j++){
htaupt->Fill(taupt[j]);
}
if (ntau >= 2){
lvectau1.SetPtEtaPhiM(taupt[0],taueta[0],tauphi[0],taumass[0]); //leading taus are the first two, that is with index 0,1
lvectau2.SetPtEtaPhiM(taupt[1],taueta[1],tauphi[1],taumass[1]);
lvecZ = lvectau1+lvectau2;
ditaumass = lvecZ.Mag(); // invariant mass
}
newtree->Fill();
}
std::cout<<"End of event loop"<<std::endl;
inputfile->Close();
//Writing output file
newFile->cd();
newFile->Write();
newFile->Close();
newFile->Delete();
std::cout<<"End of script"<<std::endl;
}
Key Points
Some help with Assignment 1
Get DELPHES
Overview
Teaching: min
Exercises: minQuestions
What is DELPHES ?
How to install DELPHES ?
Objectives
Install DELPHES
DELPHES
Delphes framework for fast detector response simulation. The simulation includes a tracking system, embedded into a magnetic field, calorimeters and a muon system. The framework is interfaced to standard file formats (e.g. Les Houches Event File or HepMC) and outputs observables such as isolated leptons, missing transverse energy and collection of jets which can be used for dedicated analyses. The simulation of the detector response takes into account the effect of magnetic field, the granularity of the calorimeters and sub-detector resolutions. Visualisation of the final state particles is also built-in using the corresponding ROOT library.
DELPHES documentation
The official webpage for DELPHES can be found here. The publication detailing DELPHES can be found at JHEP 02 (2014) 057 or arXiv:1307.6346.
Installing DELPHES
To successfully install Delphes the following prerequisite packages should be installed: ROOT and Tcl scripting language.
Download and build Delphes after sourcing ROOT:
$ wget http://cp3.irmp.ucl.ac.be/downloads/Delphes-3.5.0.tar.gz
$ tar -zxf Delphes-3.5.0.tar.gz
$ cd Delphes-3.5.0
$ make
Key Points
The official webpage for DELPHES can be found here
Introduction to DELPHES
Overview
Teaching: min
Exercises: minQuestions
What is DELPHES ?
How to use DELPHES ?
Objectives
Working with DELPHES
DELPHES
Delphes framework for fast detector response simulation. The simulation includes a tracking system, embedded into a magnetic field, calorimeters and a muon system. The framework is interfaced to standard file formats (e.g. Les Houches Event File or HepMC) and outputs observables such as isolated leptons, missing transverse energy and collection of jets which can be used for dedicated analyses. The simulation of the detector response takes into account the effect of magnetic field, the granularity of the calorimeters and sub-detector resolutions. Visualisation of the final state particles is also built-in using the corresponding ROOT library.
DELPHES documentation
The official webpage for DELPHES can be found here. The publication detailing DELPHES can be found at JHEP 02 (2014) 057 or arXiv:1307.6346.
Running DELPHES
Running Delphes can be done through the executables in the directory while passing arguments and paramteres. Eg.:
./DelphesHepMC3
But with parameters. When running Delphes without parameters or when supplying an invalid command line, the following message will be shown:
Usage: DelphesHepMC3 config_file output_file [input_file(s)]
config_file - configuration file in Tcl format,
output_file - output file in ROOT format,
input_file(s) - input file(s) in HepMC format,
with no input_file, or when input_file is -, read standard input.
Running Delphes with HepMC input files (example):
./DelphesHepMC3 cards/delphes_card_CMS.tcl output.root input.hepmc
Try using with the file ttbar.lhe shared with you.
./DelphesLHEF cards/delphes_card_CMS.tcl delphes_output.root ttbar.lhe
DELPHES produced output ROOT file
To understand the content of the ROOT file produced, a handy reference is available at the Delphes webpage linked here.
To open the file with ROOT, you need to first load the Delphes library with which the contents of the file can be read:
$ root
root[0] gSystem->Load("libDelphes");
Then you can open the Delhes output ROOT file as:
$ root
root[0] gSystem->Load("libDelphes");
root[1] TFile *_file0 = TFile::Open("delphes_output.root")
root[3] TBrowser browser
Visualising DELPHES Detector Simulation
Delphes also allows to viualise event by event detector simulation. Try it out:
$ make display
$ root -l examples/EventDisplay.C'("cards/delphes_card_CMS.tcl","delphes_output.root")'
Running DELPHES directly with PYTHIA8
For this, you need a working PYTHIA8 installation (assuming you have that from the first part of the course).
Define an environment variable for the path to your PYTHIA installation directory:
$ export PYTHIA8=path_to_PYTHIA8_installation
$ echo $PYTHIA8
Then, in your DELPHES directory, build the “DelphesPythia8” executable with the following command:
$ make HAS_PYTHIA8=true
Now, you can run a simple example for generating Pythia8 events within Delphes:
$ ./DelphesPythia8 cards/delphes_card_CMS.tcl examples/Pythia8/configNoLHE.cmnd delphes_pythia8.root
Key Points
Understanding basics of using DELPHES.
The publication detailing DELPHES can be found at JHEP 02 (2014) 057 or arXiv:1307.6346
DELPHES with Scripts
Overview
Teaching: min
Exercises: minQuestions
How to analyse DELPHES output files with scripts for ROOT?
Objectives
Learning about using DELPHES with C and Python scripts
DELPHES with basic scripts
Try to save the following script as file “DelphesScript1.C” and then run it as:
root
gSystem->Load("libDelphes");
.x DelphesScript1.C("delphes_output.root");
The script:
void DelphesScript1(const char *inputFile)
{
gSystem->Load("libDelphes");
// Create chain of root trees
TChain chain("Delphes");
chain.Add(inputFile);
// Create object of class ExRootTreeReader
ExRootTreeReader *treeReader = new ExRootTreeReader(&chain);
Long64_t numberOfEntries = treeReader->GetEntries();
// Get pointers to branches used in this analysis
TClonesArray *branchJet = treeReader->UseBranch("Jet");
TClonesArray *branchElectron = treeReader->UseBranch("Electron");
// Book histograms
TH1 *histJetPT = new TH1F("jet_pt", "jet P_{T}", 100, 0.0, 100.0);
TH1 *histMass = new TH1F("mass", "M_{inv}(e_{1}, e_{2})", 100, 40.0, 140.0);
// Loop over all events
for(Int_t entry = 0; entry < numberOfEntries; ++entry)
{
// Load selected branches with data from specified event
treeReader->ReadEntry(entry);
// If event contains at least 1 jet
if(branchJet->GetEntries() > 0)
{
// Take first jet
Jet *jet = (Jet*) branchJet->At(0);
// Plot jet transverse momentum
histJetPT->Fill(jet->PT);
// Print jet transverse momentum
cout << jet->PT << endl;
}
Electron *elec1, *elec2;
// If event contains at least 2 electrons
if(branchElectron->GetEntries() > 1)
{
// Take first two electrons
elec1 = (Electron *) branchElectron->At(0);
elec2 = (Electron *) branchElectron->At(1);
// Plot their invariant mass
histMass->Fill(((elec1->P4()) + (elec2->P4())).M());
}
}
// Show resulting histograms
histJetPT->Draw();
}
DELPHES with more advanced scripts
The following script can be run as:
root DelphesScript2.C'("delphes_output.root")'
The scripts “DelphesScript2.C” :
#include "TH1.h"
#include "TSystem.h"
#ifdef __CLING__
R__LOAD_LIBRARY(libDelphes)
#include "classes/DelphesClasses.h"
#include "external/ExRootAnalysis/ExRootTreeReader.h"
#include "external/ExRootAnalysis/ExRootResult.h"
#endif
//------------------------------------------------------------------------------
struct MyPlots
{
TH1 *fJetPT[2];
TH1 *fMissingET;
TH1 *fElectronPT;
};
//------------------------------------------------------------------------------
class ExRootResult;
class ExRootTreeReader;
//------------------------------------------------------------------------------
void BookHistograms(ExRootResult *result, MyPlots *plots)
{
THStack *stack;
TLegend *legend;
TPaveText *comment;
// book 2 histograms for PT of 1st and 2nd leading jets
plots->fJetPT[0] = result->AddHist1D(
"jet_pt_0", "leading jet P_{T}",
"jet P_{T}, GeV/c", "number of jets",
50, 0.0, 100.0);
plots->fJetPT[1] = result->AddHist1D(
"jet_pt_1", "2nd leading jet P_{T}",
"jet P_{T}, GeV/c", "number of jets",
50, 0.0, 100.0);
plots->fJetPT[0]->SetLineColor(kRed);
plots->fJetPT[1]->SetLineColor(kBlue);
// book 1 stack of 2 histograms
stack = result->AddHistStack("jet_pt_all", "1st and 2nd jets P_{T}");
stack->Add(plots->fJetPT[0]);
stack->Add(plots->fJetPT[1]);
// book legend for stack of 2 histograms
legend = result->AddLegend(0.72, 0.86, 0.98, 0.98);
legend->AddEntry(plots->fJetPT[0], "leading jet", "l");
legend->AddEntry(plots->fJetPT[1], "second jet", "l");
// attach legend to stack (legend will be printed over stack in .eps file)
result->Attach(stack, legend);
// book more histograms
plots->fElectronPT = result->AddHist1D(
"electron_pt", "electron P_{T}",
"electron P_{T}, GeV/c", "number of electrons",
50, 0.0, 100.0);
plots->fMissingET = result->AddHist1D(
"missing_et", "Missing E_{T}",
"Missing E_{T}, GeV", "number of events",
60, 0.0, 30.0);
// book general comment
comment = result->AddComment(0.64, 0.86, 0.98, 0.98);
comment->AddText("demonstration plot");
comment->AddText("produced by Example2.C");
// attach comment to single histograms
result->Attach(plots->fJetPT[0], comment);
result->Attach(plots->fJetPT[1], comment);
result->Attach(plots->fElectronPT, comment);
// show histogram statisics for MissingET
plots->fMissingET->SetStats();
}
//------------------------------------------------------------------------------
void AnalyseEvents(ExRootTreeReader *treeReader, MyPlots *plots)
{
TClonesArray *branchJet = treeReader->UseBranch("Jet");
TClonesArray *branchElectron = treeReader->UseBranch("Electron");
TClonesArray *branchMissingET = treeReader->UseBranch("MissingET");
Long64_t allEntries = treeReader->GetEntries();
cout << "** Chain contains " << allEntries << " events" << endl;
Jet *jet[2];
MissingET *met;
Electron *electron;
Long64_t entry;
Int_t i;
// Loop over all events
for(entry = 0; entry < allEntries; ++entry)
{
// Load selected branches with data from specified event
treeReader->ReadEntry(entry);
// Analyse two leading jets
if(branchJet->GetEntriesFast() >= 2)
{
jet[0] = (Jet*) branchJet->At(0);
jet[1] = (Jet*) branchJet->At(1);
plots->fJetPT[0]->Fill(jet[0]->PT);
plots->fJetPT[1]->Fill(jet[1]->PT);
}
// Analyse missing ET
if(branchMissingET->GetEntriesFast() > 0)
{
met = (MissingET*) branchMissingET->At(0);
plots->fMissingET->Fill(met->MET);
}
// Loop over all electrons in event
for(i = 0; i < branchElectron->GetEntriesFast(); ++i)
{
electron = (Electron*) branchElectron->At(i);
plots->fElectronPT->Fill(electron->PT);
}
}
}
//------------------------------------------------------------------------------
void PrintHistograms(ExRootResult *result, MyPlots *plots)
{
result->Print("png");
}
//------------------------------------------------------------------------------
void DelphesScript2(const char *inputFile)
{
gSystem->Load("libDelphes");
TChain *chain = new TChain("Delphes");
chain->Add(inputFile);
ExRootTreeReader *treeReader = new ExRootTreeReader(chain);
ExRootResult *result = new ExRootResult();
MyPlots *plots = new MyPlots;
BookHistograms(result, plots);
AnalyseEvents(treeReader, plots);
PrintHistograms(result, plots);
result->Write("results.root");
cout << "** Exiting..." << endl;
delete plots;
delete result;
delete treeReader;
delete chain;
}
DELPHES with python scripts
The following script can be run as:
python DelphesPythonScript.py delphes_output.root
The script “DelphesPythonScript.py” :
#!/usr/bin/env python
import sys
import ROOT
try:
input = raw_input
except:
pass
if len(sys.argv) < 2:
print(" Usage: Example1.py input_file")
sys.exit(1)
ROOT.gSystem.Load("libDelphes")
try:
ROOT.gInterpreter.Declare('#include "classes/DelphesClasses.h"')
ROOT.gInterpreter.Declare('#include "external/ExRootAnalysis/ExRootTreeReader.h"')
except:
pass
inputFile = sys.argv[1]
# Create chain of root trees
chain = ROOT.TChain("Delphes")
chain.Add(inputFile)
# Create object of class ExRootTreeReader
treeReader = ROOT.ExRootTreeReader(chain)
numberOfEntries = treeReader.GetEntries()
# Get pointers to branches used in this analysis
branchJet = treeReader.UseBranch("Jet")
branchElectron = treeReader.UseBranch("Electron")
# Book histograms
histJetPT = ROOT.TH1F("jet_pt", "jet P_{T}", 100, 0.0, 100.0)
histMass = ROOT.TH1F("mass", "M_{inv}(e_{1}, e_{2})", 100, 40.0, 140.0)
# Loop over all events
for entry in range(0, numberOfEntries):
# Load selected branches with data from specified event
treeReader.ReadEntry(entry)
# If event contains at least 1 jet
if branchJet.GetEntries() > 0:
# Take first jet
jet = branchJet.At(0)
# Plot jet transverse momentum
histJetPT.Fill(jet.PT)
# Print jet transverse momentum
print(jet.PT)
# If event contains at least 2 electrons
if branchElectron.GetEntries() > 1:
# Take first two electrons
elec1 = branchElectron.At(0)
elec2 = branchElectron.At(1)
# Plot their invariant mass
histMass.Fill(((elec1.P4()) + (elec2.P4())).M())
# Show resulting histograms
histJetPT.Draw()
input("Press Enter to continue...")
More complex Python code can be written based on a separate analysis package, deteailed at DelphesAnalysis.
Key Points
Learning about using DELPHES with C and Python scripts
More complex Python code can be written based on a separate analysis package, deteailed at DelphesAnalysis.